
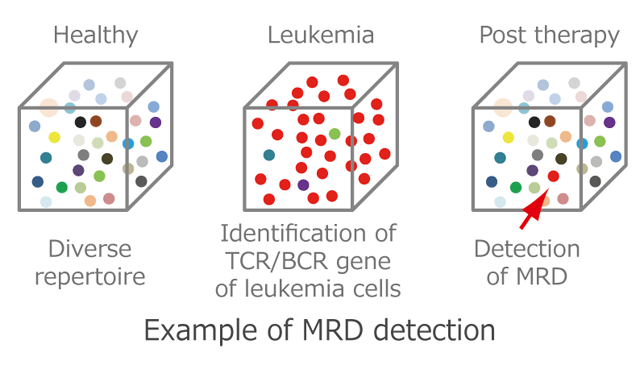
To date, several pipelines that perform both BcR IG/TR NGS immunogenetics has applications in both diagnostics (e.g., assessment of clonality in samples investigated for a possible lymphoproliferation or detection of minimal residual disease in patients with lymphoid malignancies) and research.
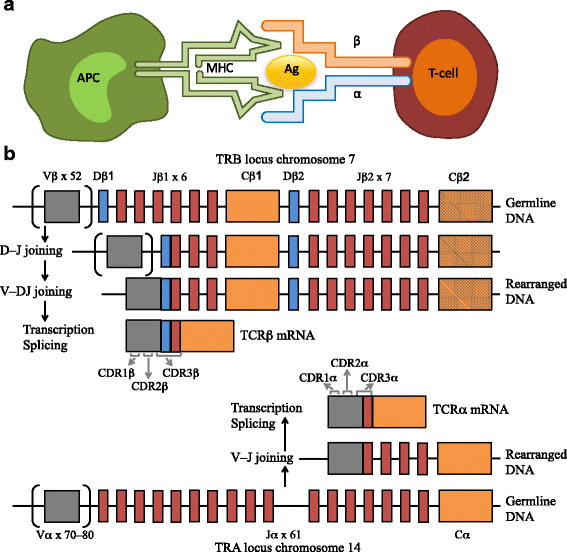
) gene repertoires using next-generation sequencing (NGS) technologies advanced our understanding of various clinical conditionsĪnd biological processes, extending from infections, vaccination, autoimmunity, to malignancy. Profiling the B-cell receptor immunoglobulin (BcR IG) and T-cell receptor (TR Relevant to mention, TRIP enables a high level of customization through the integration of various options in key aspects of the analysis, such as clonotype definition and computation, hence allowing for flexibility without compromising on accuracy. Subsequently, it provides in-depth analysis of antigen receptor gene rearrangements, including (a) clonality assessment (b) extraction of variable (V), diversity (D), and joining (J) gene repertoires (c) CDR3 characterization at both the nucleotide and amino acid level and (d) somatic hypermutation analysis, in the case of immunoglobulin gene rearrangements.
#TCR REPERTOIRE IMGT SERIES#
TRIP has the potential to perform robust immunoprofiling analysis through the extraction and processing of the IMGT/HighV-Quest output, via a series of functions, ensuring the analysis of high-quality, biologically relevant data through a multilevel process of data filtering. Several bioinformatics workflows have been developed to this end, including the T-cell receptor/immunoglobulin profiler (TRIP), a web application implemented in R shiny, specifically designed for the purposes of comprehensive repertoire analysis, which is the focus of this chapter. The IGKV genes of the distal duplicated V-CLUSTER are designated by the same number as the corresponding genes in the proximal V-CLUSTER, with the letter D added.The study of antigen receptor gene repertoires using next-generation sequencing (NGS) technologies has disclosed an unprecedented depth of complexity, requiring novel computational and analytical solutions. IGKV gene names are designated by a number for the subgroup, followed by a dash and a number for the localization from 3' to 5' in the locus. clan III: IGKV5 and IGKV7 subgroup genes.clan II: IGKV2, IGKV3, IGKV4 and IGKV6 subgroup genes.The 7 human (Homo sapiens) IGKV subgroups belong to three clans accession number to get the corresponding IMGT/LIGM-DB entry.IMGT allele name to see the corresponding Alignments of alleles,.IMGT gene name to get the corresponding IMGT/GENE-DB entry,.Refers to genomic DNA, but not known as being germline or rearranged. Number refers to rearranged genomic DNA or cDNA and the corresponding germlineįunctionality is shown between brackets, and, when the accession number When the data have been confirmed by several studies.įunctionality is shown between parentheses, (F) and (P), when the accession Arbitrarily that information is shown on the first line of each gene Transcribed (T) and/or translated into protein "+" or "-" indicates if the gene sequences have been found (+) or not been found (-)


 0 kommentar(er)
0 kommentar(er)
